Difference between revisions of "Goals"
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | Our | + | Our aims support the development and evaluation of novel methods for cancer deep phenotype extraction: |
| − | '''Specific Aim 1''': Develop methods for extracting phenotypic profiles | + | '''Specific Aim 1''': Develop methods for extracting phenotypic profiles |
'''Specific Aim 2''': Extract gene/protein mentions and their variants from the clinical narrative | '''Specific Aim 2''': Extract gene/protein mentions and their variants from the clinical narrative | ||
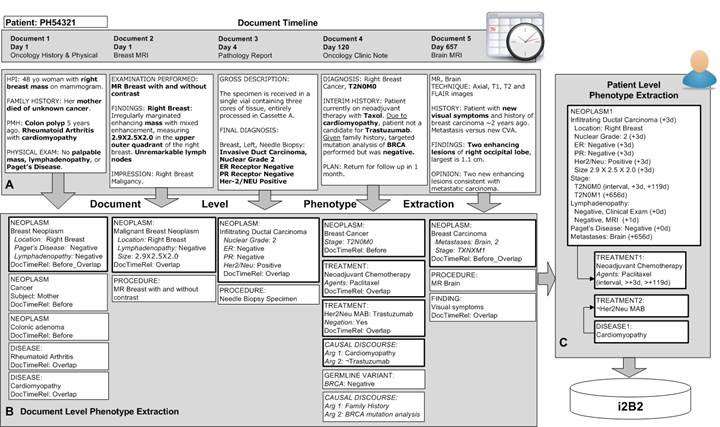
| − | '''Specific Aim 3''': Create longitudinal representation of disease process and its resolution | + | '''Specific Aim 3''': Create longitudinal representation of disease process and its resolution |
| − | [[File:Aim 3.jpg| | + | [[File:Aim 3.jpg| center]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
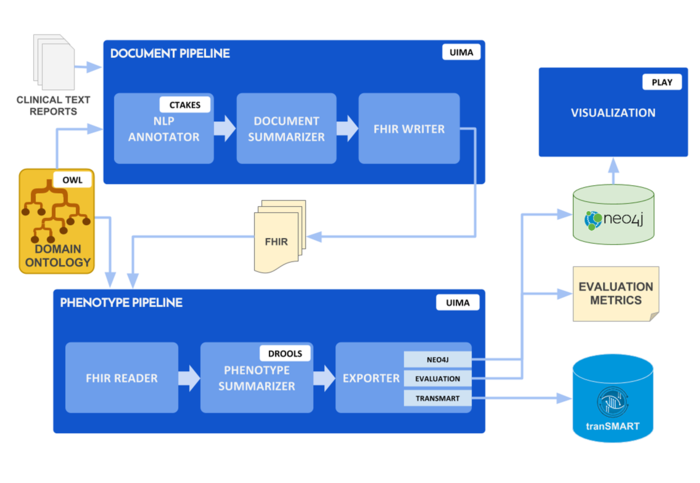
'''Specific Aim 5''': Design and implement a computational platform for deep phenotype discovery and analytics for translational investigators, including integrative visual analytics. | '''Specific Aim 5''': Design and implement a computational platform for deep phenotype discovery and analytics for translational investigators, including integrative visual analytics. | ||
| − | [[File:Architecture overview.png| center | | + | [[File:Architecture overview.png| center | 700px]] |
'''Specific Aim 6''': Advance translational research in driving cancer biology research projects in breast cancer, ovarian cancer, and melanoma. Include research community throughout the design of the platform and its evaluation. Disseminate freely available software. | '''Specific Aim 6''': Advance translational research in driving cancer biology research projects in breast cancer, ovarian cancer, and melanoma. Include research community throughout the design of the platform and its evaluation. Disseminate freely available software. | ||
'''''Impact:''''' The proposed work will produce novel methods for extracting detailed phenotype information directly from the EMR, the major source of such data for patients with cancer. Extracted phenotypes will be used in three ongoing translational studies with a precision medicine focus. Dissemination of the software will enhance the ability of cancer researchers to abstract meaningful clinical data for translational research. If successful, systematic capture and representation of these phenotypes from EMR data could later be used to drive clinical genomic decision support. | '''''Impact:''''' The proposed work will produce novel methods for extracting detailed phenotype information directly from the EMR, the major source of such data for patients with cancer. Extracted phenotypes will be used in three ongoing translational studies with a precision medicine focus. Dissemination of the software will enhance the ability of cancer researchers to abstract meaningful clinical data for translational research. If successful, systematic capture and representation of these phenotypes from EMR data could later be used to drive clinical genomic decision support. | ||
Latest revision as of 15:12, 9 April 2019
Our aims support the development and evaluation of novel methods for cancer deep phenotype extraction:
Specific Aim 1: Develop methods for extracting phenotypic profiles
Specific Aim 2: Extract gene/protein mentions and their variants from the clinical narrative
Specific Aim 3: Create longitudinal representation of disease process and its resolution
Specific Aim 5: Design and implement a computational platform for deep phenotype discovery and analytics for translational investigators, including integrative visual analytics.
Specific Aim 6: Advance translational research in driving cancer biology research projects in breast cancer, ovarian cancer, and melanoma. Include research community throughout the design of the platform and its evaluation. Disseminate freely available software.
Impact: The proposed work will produce novel methods for extracting detailed phenotype information directly from the EMR, the major source of such data for patients with cancer. Extracted phenotypes will be used in three ongoing translational studies with a precision medicine focus. Dissemination of the software will enhance the ability of cancer researchers to abstract meaningful clinical data for translational research. If successful, systematic capture and representation of these phenotypes from EMR data could later be used to drive clinical genomic decision support.